2015 AMIA Joint Summits on Translational Science
The American Medical Informatics Association (AMIA) hosted the 2015 Joint Summits on Translational Science in San Fransisco. The first half of the conference featured work on translational bioinformatics, and the second half featured work on clinical research informatics.
ResearchMaps poster
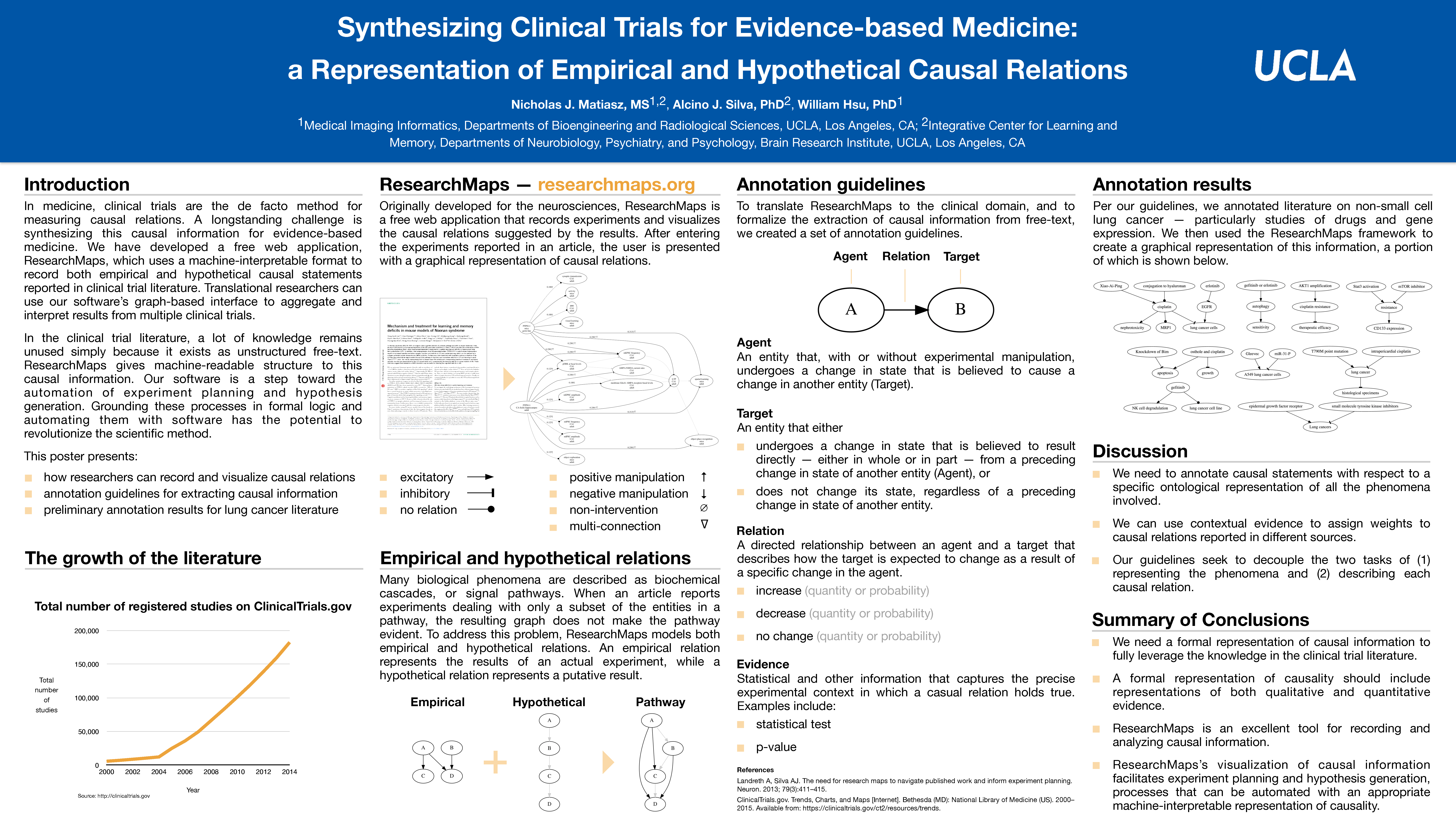
During the second half of this conference, I presented a poster titled “Synthesizing Clinical Trials for Evidence-based Medicine: a Representation of Empirical and Hypothetical Causal Relations.” Building off of the work I presented at the 13th Molecular and Cellular Cognition Society Meeting, this poster presented our efforts to use ResearchMaps to represent causal relations in the clinical trial literature.
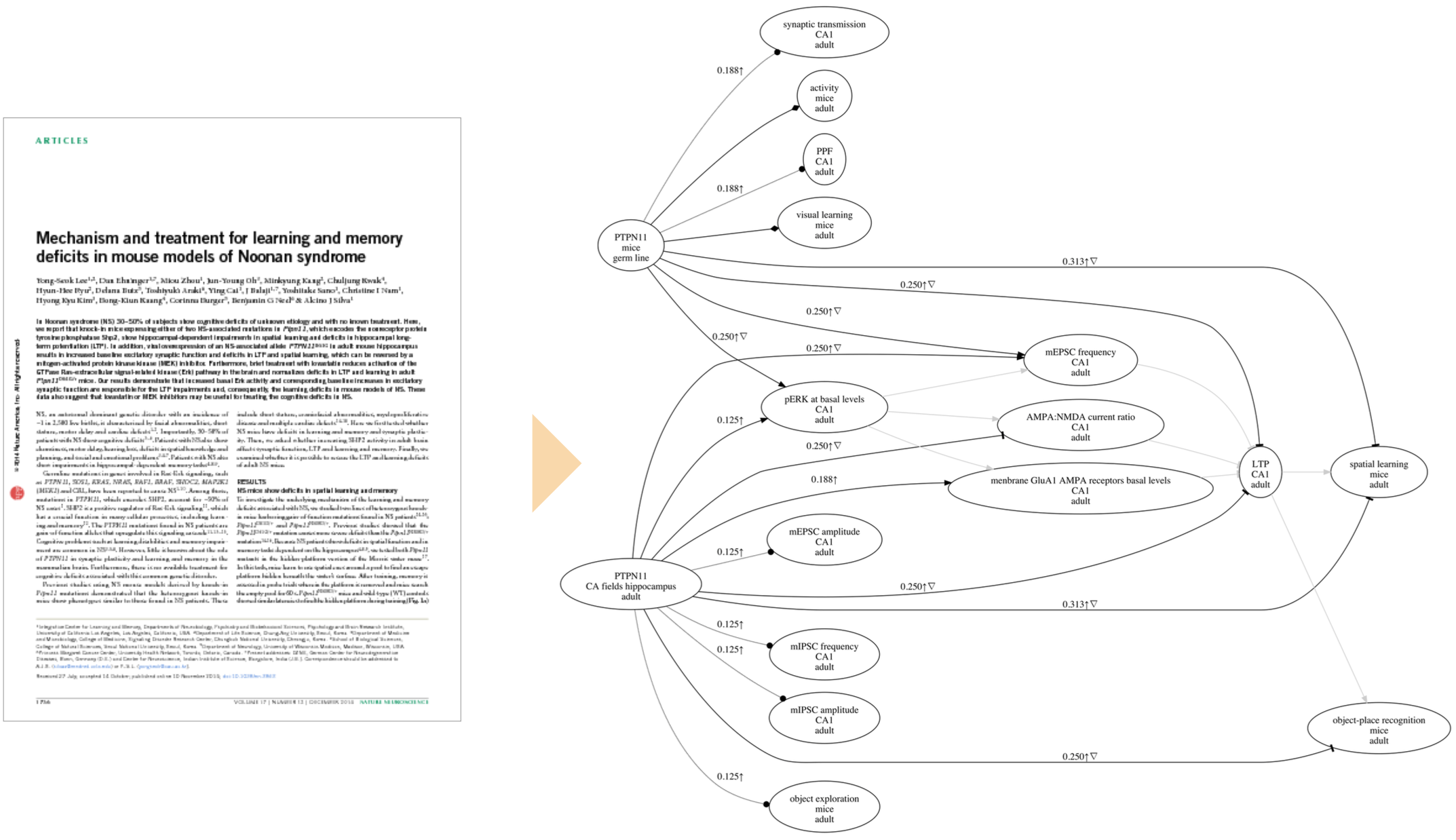
Our poster also reported one of our newest features in ResearchMaps: the ability to represent both empirical and hypothetical causal relations in a single causal graph. We developed this feature to address an obvious fact: When we read a research paper, we don’t only consider the information presented in the text; we also use our domain knowledge to contextualize, interpret, and structure the information.
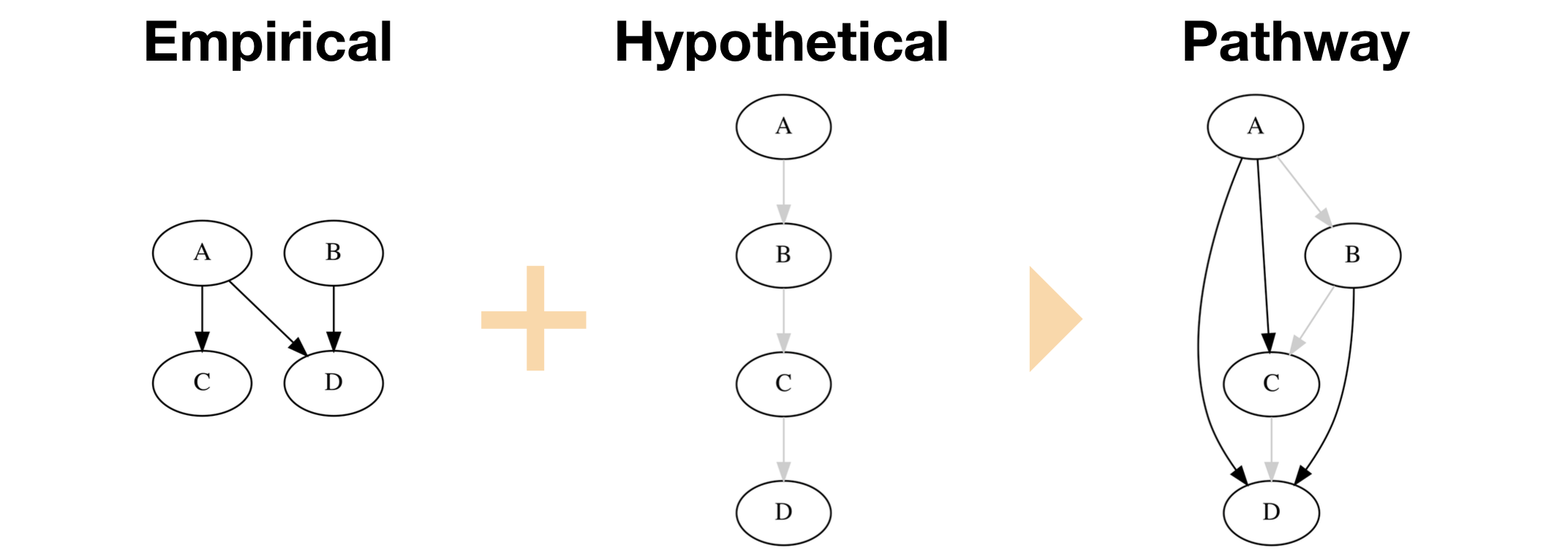
To see why this feature is useful, consider a causal graph that models a biochemical cascade, or signal pathway. We’ll model this pathway as A -> B -> C -> D. Now consider a research paper with experiments that deal only with a subset of the entities in the pathway—for example, experiments that test the relations A -> C, A -> D, and B -> D. The resulting graph of these experiments does not make the pathway evident. However, if you believe that the A -> B -> C -> D pathway exists, you’ll use this information to interpret the results of the reported experiments. To visualize this contextual knowledge, ResearchMaps allows you to model both the empirical causal relations—those for which a publication offers supporting data—as well as hypothetical relations—putative relations for which a publication has no supporting data.
References
For a sampling of the people who attended this conference and the publications, ideas, etc. that were discussed, I’ve compiled a short list of references. This list is not meant to be exhaustive in any way; it simply reflects who and what caught my attention.
People
- Russ B. Altman, MD, PhD
- Douglas S. Bell, MD, PhD, FACMI
- Nigam H. Shah, MBBS, PhD
- Ida Sim, PhD, MD
- Paul Wicks, PhD
Publications
- D. Estrin and I. Sim, “Open mhealth architecture: an engine for health care innovation,” Science, vol. 330, no. 6005, pp. 759–760, 2010.
- S. K. Gire, A. Goba, K. G. Andersen, R. S. Sealfon, D. J. Park, L. Kanneh, S. Jalloh, M. Momoh, M. Fullah, G. Dudas et al., “Genomic surveillance elucidates ebola virus origin and transmission during the 2014 outbreak,” Science, vol. 345, no. 6202, pp. 1369–1372, 2014.
- K. Jensen, G. Panagiotou, and I. Kouskoumvekaki, “Integrated text mining and chemoinformatics analysis associates diet to health benefit at molecular level,” PLoS computational biology, vol. 10, no. 1, p. e1003432, 2014.
- S. Mukherjee, The emperor of all maladies: a biography of cancer. Simon and Schuster, 2010.
- G. M. Weber, K. D. Mandl, and I. S. Kohane, “Finding the missing link for big biomedical data,” JAMA, vol. 311, no. 24, pp. 2479–2480, 2014.
- A. Wilcox and E. Holve, “Sustaining the effective use of health care data: A message from the editors,” eGEMs (Generating Evidence & Methods to improve patient outcomes), vol. 2, no. 2, p. 9, 2014.
Organizations, software, etc.
- BioPortal
- ConceptQL
- GeoJSON
- Mobile Sensor Data-to-Knowledge
- Observational Health Data Sciences and Informatics (OHDSI)
- Observational Medical Outcomes Partnership (OMOP) Common Data Model
- Open mHealth
- PatientsLikeMe
- OrientDB
- RexDB
⁂
Subscribe to my mailing list if you’d like to read more.